Objective
Structure
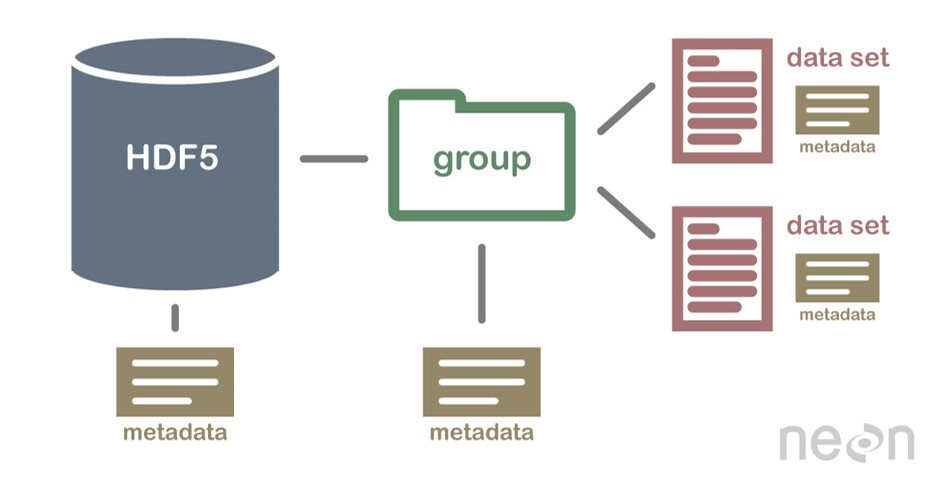
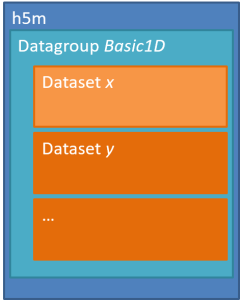
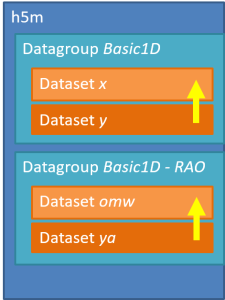
An h5m file is organised around a two level structure:
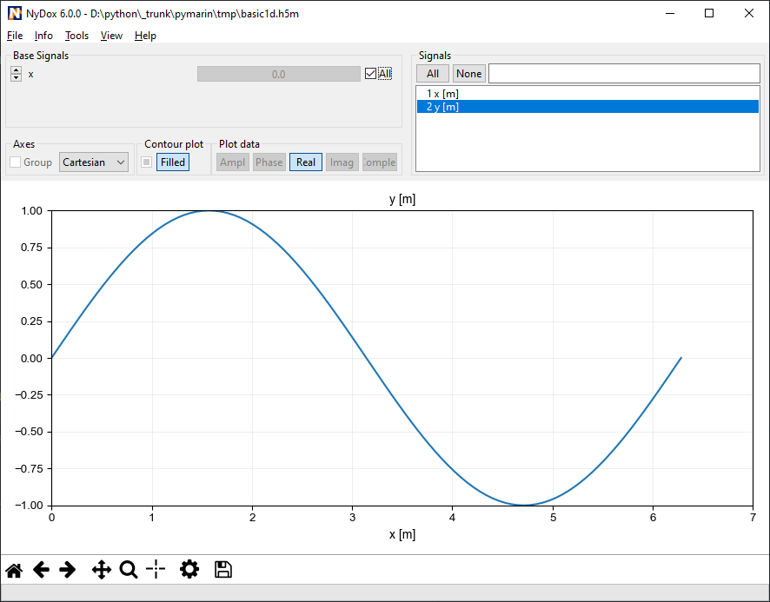
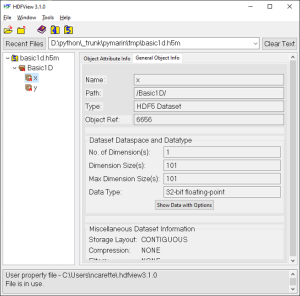
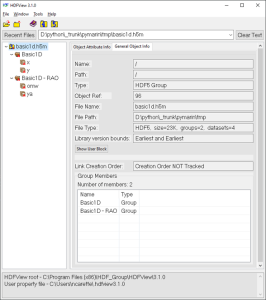
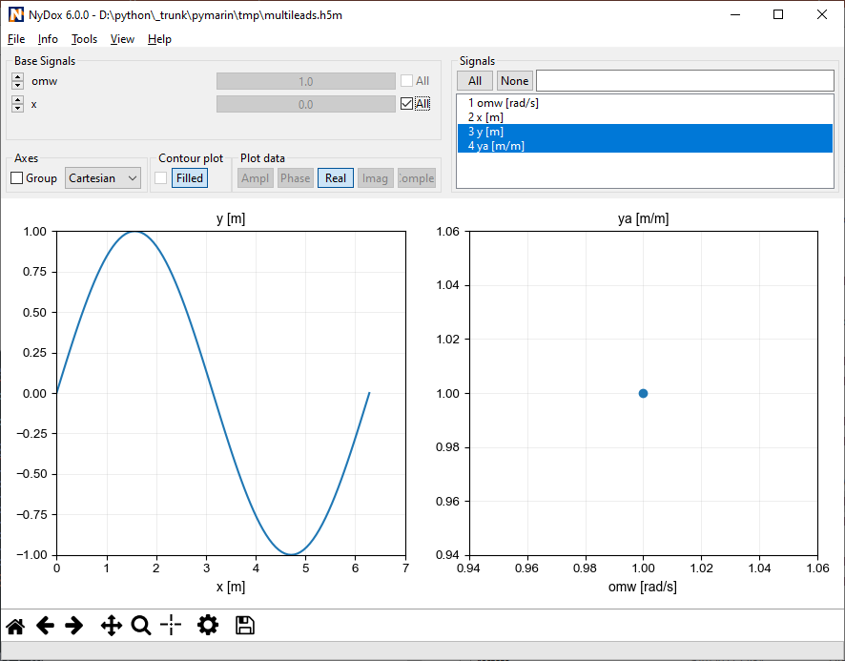
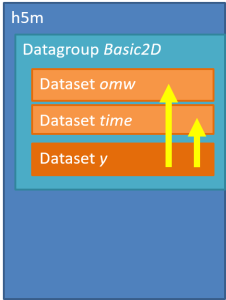
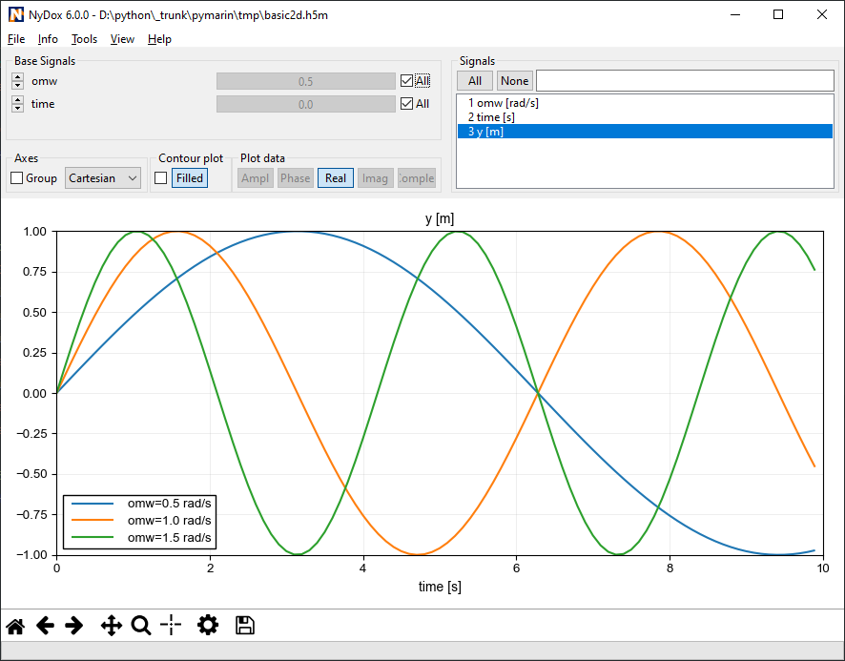
There may be as many groups and data sets as necessary, but not nested structure is allowed (v0.1). A set of metadata can be stored at each level as attributes. The h5m specification describes in detail which attributes are expected and which are required. The data set contains the data of one signal. Relation between the signals can be given, signals that are independent are base or lead signals. For example, a group can be created with one lead signal x, and a series of dependent signals. A schematic representation, with a simple x-y plot example and view in HDFView is given here under. In this case a simple y=sin(x) data was stored in an h5m file.
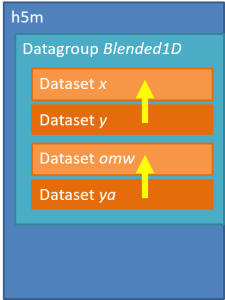
Several groups can be stored in one file, for instance to store measured and analysed data in the same file. For example, we can store the amplitude and frequency data of the y=sin(x) in the previous example. The signals in the second group are simple 1 value arrays, but this example could be easily expanded.
However, it is allowed to store all the signals in the same group.
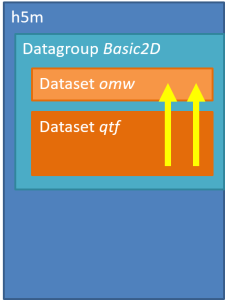
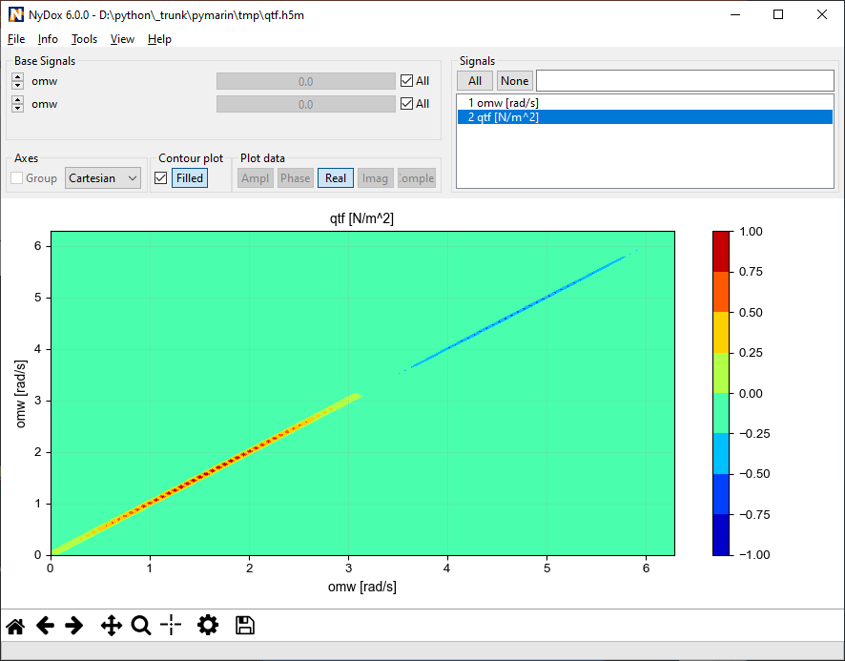
It is also possible to create N-D data by specifying multiple lead signals to one dependent signal. The order of the leads has a direct influence on the shape of the dependent signal. A signal may even depend multiple times of the same lead signal.
Group and data types
H5M has three types of groups, the type is given as a string in the attributes:
- General: for any kind of data
- Time: the group must contain at least one time base and all dependent signals must have as last dependence the time. Note that some workflows at MARIN (pymarin, SHARK) expect one unique time base per group to simplify processing.
- Frequency: similar to Time, but with a frequency base.
Data sets may have any data type allowed by hdf5. Signals may contain thus float, but also integer, boolean or string data. Note that hdf5 does not provide a type for complex numbers. MARIN follows h5py's default approach here using compound numbers with 'r' and 'i' data.